PAcAlCI
PAcAlCI (Prediction of Accuracy in Alignments based on Computational Intelligence) is novel intelligent algorithm based on least square support vector machine (LS-SVM) to predict how accurately ten different MSA tools could align a particular set of sequences.
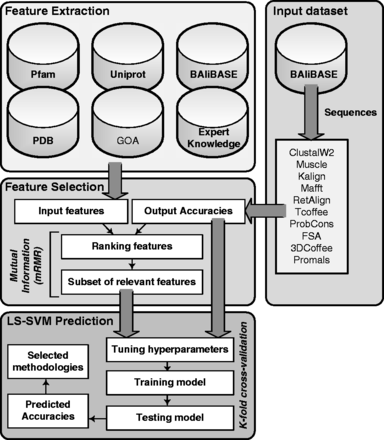
PAcAlCI is composed by four independent modules: (i) Input Dataset, (ii) Feature Extraction, (iii) Feature Selection and (iv) LS-SVM Prediction. The full PAcAlCI system was implemented with Matlab ® (Version R2010b). The MATLAB function PAcAlCI.m predicts the accuracy of the 10 alignment methodologies. The input data must be a FASTA file including sequences to align.
This function applies the LS-SVM model and confidence intervals as described in [LINK].
This function also requires the installation of the Matlab toolbox called LSSVMLab. This toolbox can be found in:
http://www.esat.kuleuven.be/sista/lssvmlab/
Usage
% Run PAcAlCI [accuracies, methods] = PAcAlCI('BB50010.tfa');
% PAcAlCI returns two variables:
% 1) accuracies -> The 10 predicted accuracies for each alignment
% methodology. For example, the first value returns the predicted accuracy
% for ClustalW:
accuracies(1)
% 2) methods -> Methodologies which have been selected as suitable for the % sequences (array).
methods
Download
Please, cite this work:
Ortuño, F.M., Valenzuela, O., Pomares, H., Rojas, F., Florido, J.P., Urquiza, J.M., Rojas, I.: Predicting the accuracy of multiple sequence alignment algorithms by using computational intelligent techniques. Nucleic Acids Research 41, e26 (2013).[LINK]