MO-SAStrE
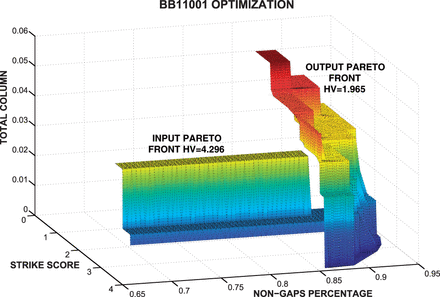
MO-SAStrE (Multiobjective Optimization for Sequence Alignments based on Structural Evaluations) proposes a multiobjective genetic approach that takes advantage of three different objectives: STRIKE score (based on structural information), totally conserved (TC) columns and percentage of non-gaps. Additionally, this algorithm also applies a novel codification of individuals as well as efficient mutation and crossover operators. This algorithm is implemented through the well-known multiobjective non-dominated sorting genetic algorithm (NSGA-II) approach.
MO-SAStrE aims to improve the quality of other alignments previously built from other alignments tools focusing on these specific three objectives. Therefore, alignments in MO-SAStrE are progressively assembled including different sections from initial alignments (crossover) and gaps shifts (mutation).
This program has been developed with Matlab ® (R2010b). Specifically, the MATLAB function MOSAStrE.m run the multibojective optimization of some input alignments. The input data must be several FASTA or GCG files including those alignments to optimize. This function applies the NSGA-II evolutionary algorithm from the Matlab toolbox for multiobjective genetic algorithms.
Download
Please, cite this work:
Ortuño, F.M., Valenzuela, O., Rojas, F., Pomares, H., Florido, J.P., Urquiza, J.M., Rojas, I.: Optimizing multiple sequence alignments using a genetic algorithm based on three objectives: structural information, non-gaps percentage and totally conserved columns. Bioinformatics 29, 2112-2121 (2013).[LINK]
Contact:
fortuno@ugr.es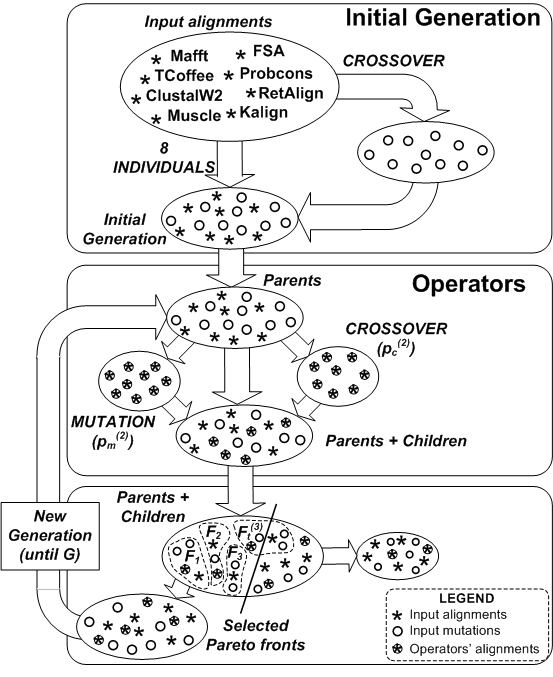
Usage
% Run PAcAlCI [accuracies, methods] = PAcAlCI('BB50010.tfa');
% Build array with paths of alignment files. We have applied alignments for eight known methodologies:
% ClustalW, Muscle, TCoffee, ProbCons, FSA, RetAlign, Mafft and Kalign.
alignments = [{'./Example/BB20020_clustalw.msf'}; {'./Example/BB20020_muscle.msf'};
{'./Example/BB20020_tcoffee.msf'}; {'./Example/BB20020_probcons.msf'};...
{'./Example/BB20020_fsa.msf'}; {'./Example/BB20020_retalign.msf'};...
{'./Example/BB20020_mafft.msf'}; {'./Example/BB20020_kalign.msf'}];
% Run MO-SAStrE using contact file (optimal run): opt_alignment = MOSAStrE(alignments,'CONTACTS',{'./Example/BB20020_contacts.txt'});
% The procedure can also run without such information but it then needs additional information: % 1) Calculate contacts providing one structure (slower). opt_alignment = MOSAStrE(alignments,'STRUCTURES',{'./Example/1mrj_.pdb'});
% 2) Calculate contacts providing all the structures (slower).
structures = [{'./Example/1mrj_.pdb'}; {'./Example/1apg_A.pdb'}; {'./Example/1abr_A.pdb'};...
{'./Example/1qi7_A.pdb'}; {'./Example/1apa_.pdb'}; {'./Example/1dm0_A.pdb'}];...
opt_alignment = MOSAStrE(alignments,'STRUCTURES',structures);
% 3) Download structures and calculate contacts from provided structures (slower).
opt_alignment = MOSAStrE(alignments);
% The optimized alignment can be saved in a file:
multialignwrite('./Example/Optimized_Alignment.msf',opt_alignment);